|
|
Topology-Preserving Selection and Clustering (TPSC)
GO TO ➢ [ Summary · Vector Space Model · SOM · SVD ] ➢ [ Hybrid SOM-SVD · Two-Phase Clustering ] ➢ [ HOWTO ] ➢ [ Citations ]
Two-Phase Clustering
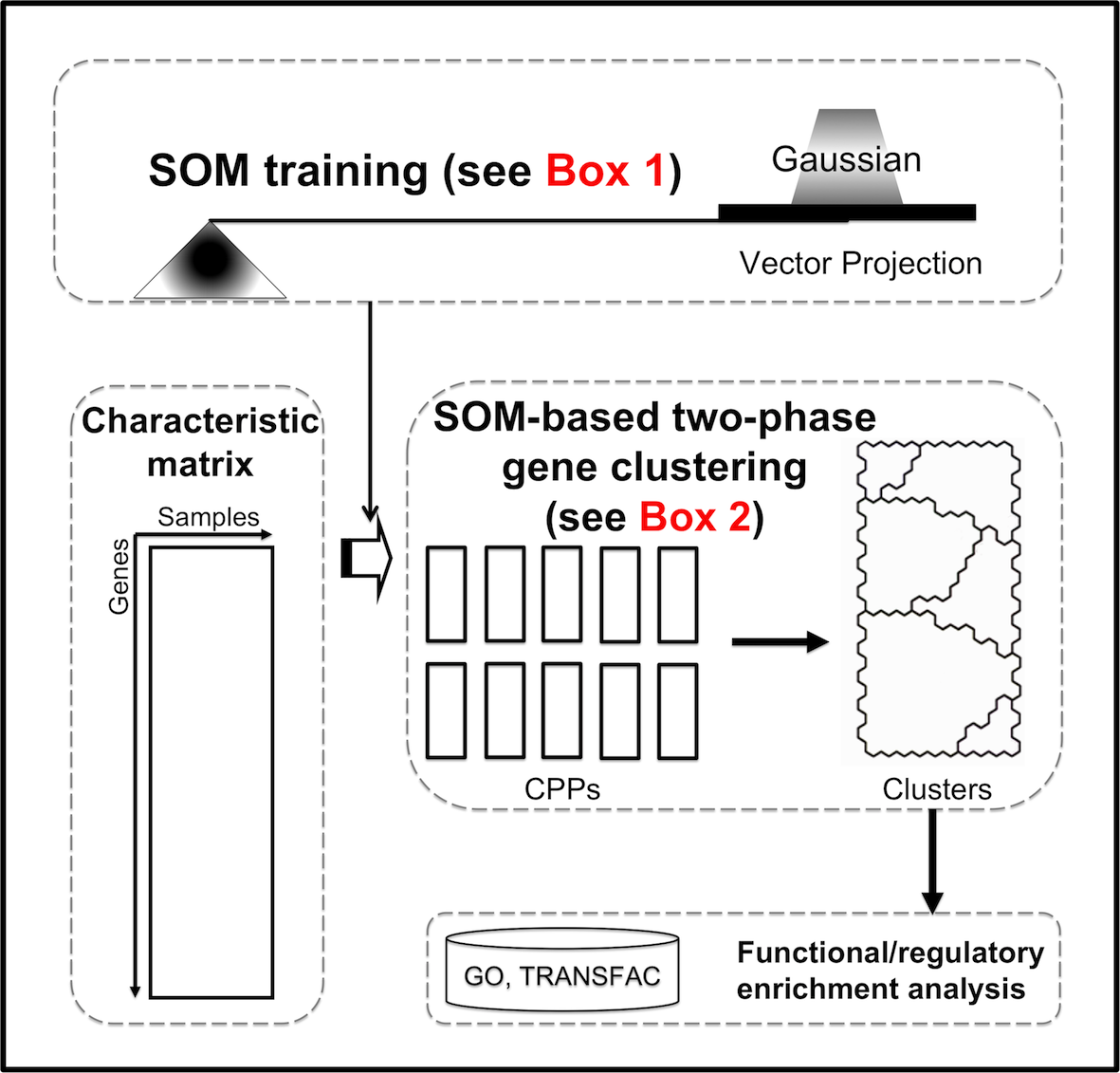
Two-phase clustering is SOM-based, allowing the topology-preserving identification of gene clusters. In the first phase, SOM training with Gaussian neighborhood kernel is applied to better preserve the topology (vector projection, VP), which can be visualized in component plane presentations (CPPs). In the second phase, distance matrix-based clustering of SOM is performed to obtain clusters without a priori assumption of data structure. Figure 1 summarizes the procedures how to implement this approach.
|
|
Figure 1. Flowchart of procedures to topology-preserving gene clustering.
|
SOM training with Gaussian kernel function The tabulated gene expression characteristic matrix, an expression matrix of genes (rows) against different experimental samples (columns) which are selected by hybrid SOM-SVD, are trained by SOM with the Gaussian kernel (see Box 1) to better preserve the topology of the data.
|
Box 1 SOM training with Gaussian kernel function.
|

|
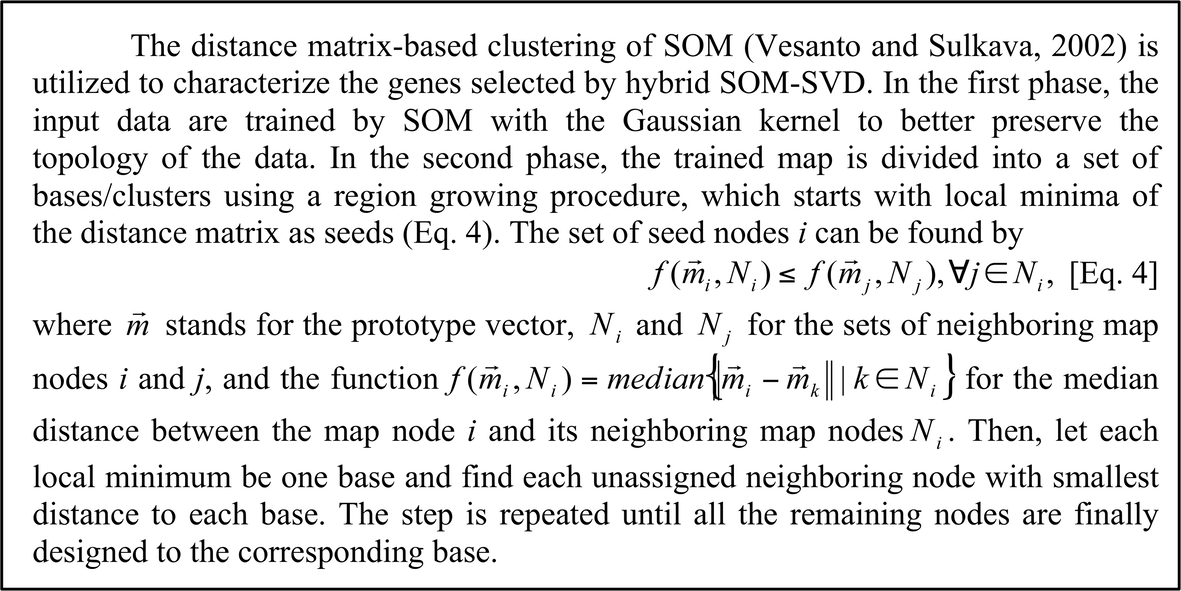
Distance matrix-based clustering of SOM The trained map is then divided into a set of clusters using a region growing procedure (see Box 2).
|
Box 2 Distance matrix-based clustering of SOM.
|
|
|
|