|
|
Topology-Preserving Selection and Clustering (TPSC)
GO TO ➢ [ Summary · Vector Space Model · SOM · SVD ] ➢ [ Hybrid SOM-SVD · Two-Phase Clustering ] ➢ [ HOWTO ] ➢ [ Citations ]
Hybrid SOM-SVD
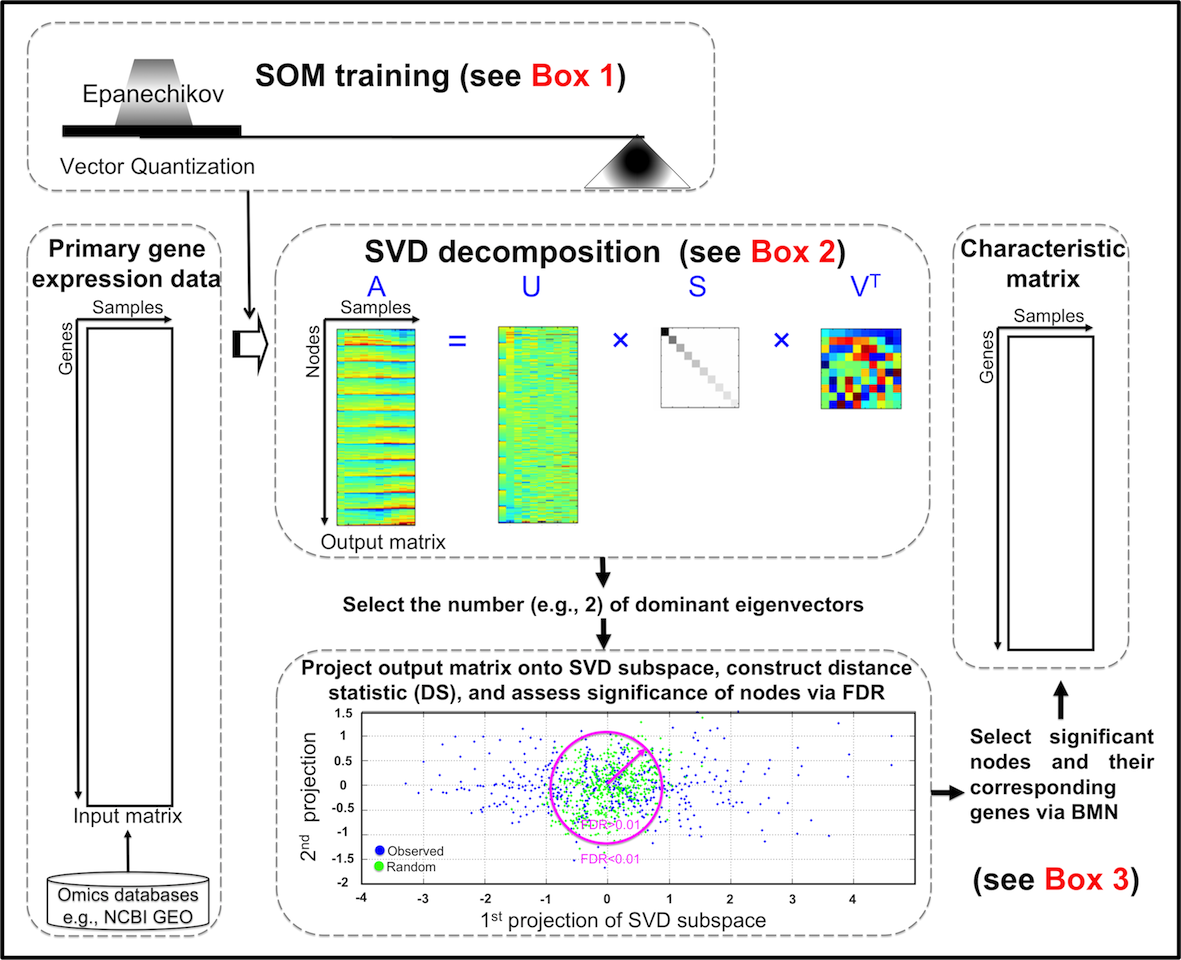
The motivations behind are to integrate the functionalities of SOM for data pre-processing and SVD for pattern recognition, thus allowing a topological-preserved selection of regulated genes based on sound foundations of statistical inference. Figure 1 summarizes the procedures how to implement this approach.
|
|
Figure 1. Flowchart of procedures to topology-preserving gene selection.
|
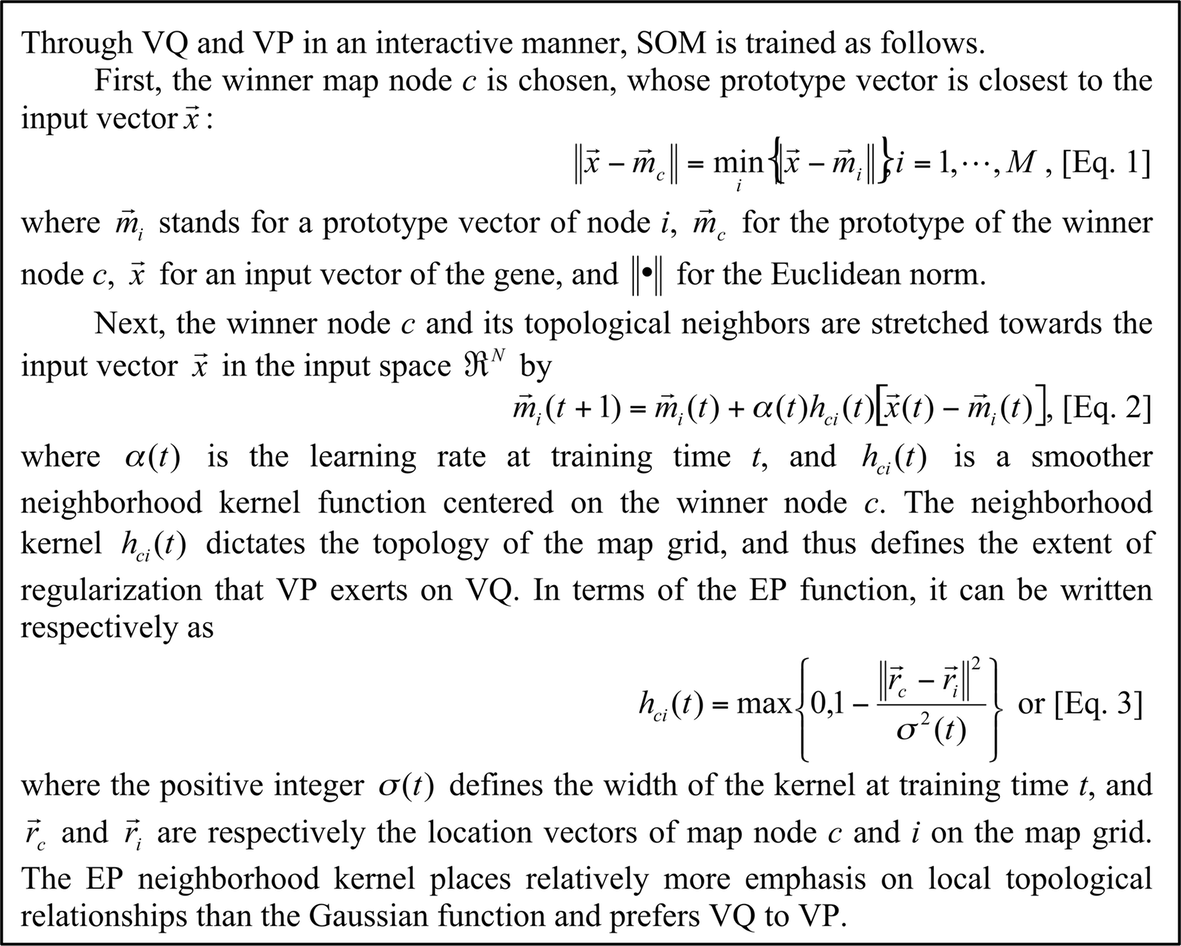
SOM training with Epanechikov (EP) kernel function The tabulated gene expression matrix (as input matrix), usually an expression matrix of genes (rows) against different experimental samples (columns), is subjected to non-linear transformation using SOM with EP neighborhood kernel, with emphasis on vector quantization (VQ) (see Box 1).
|
Box 1 SOM training with Epanechikov (EP) kernel function.
|
|
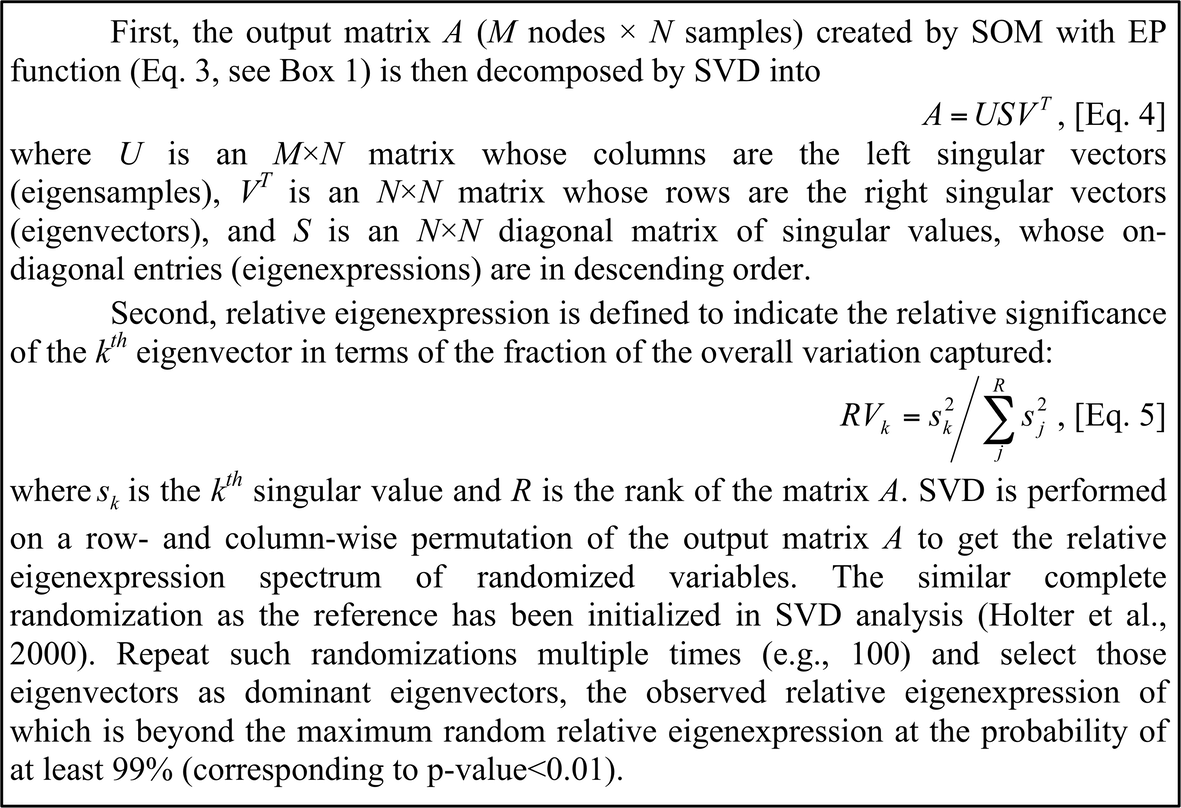
Hybrid SOM-SVD for topology-preserving gene selection The resultant output matrix (i.e., nodes in rows × samples in columns) serves as an intermediate format for pattern recognition by SVD, which is followed by dominant eigenvector selection (see Box 2). After that, it is sequentially subjected to SVD subspace projection and distance statistics construction, significant node assessment using the false discovery rate (FDR) procedure for multiple hypothesis testing, and finally selection of significant nodes and their corresponding genes defined by the best-matching node (BMN) (see Box 3). Those primary gene expression data selected through hybrid SOM-SVD form the characteristic matrix, which remains for further analysis.
|
Box 2 Decomposition by SVD, and selection of the dominant eigenvectors.
|
|
|
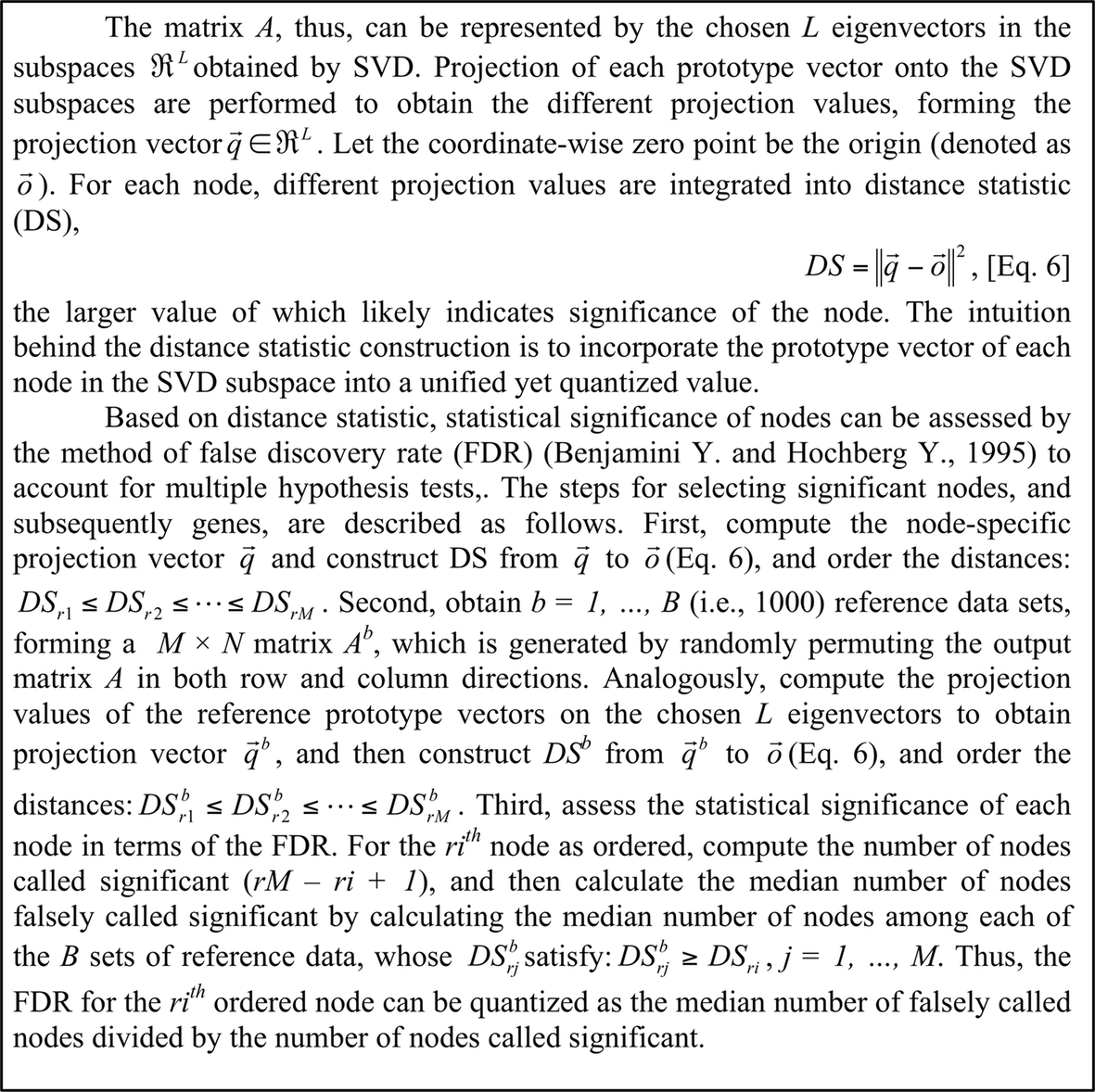
Box 3 SVD subspace projection, distance statistics construction, and assessment of significant nodes.
|
|
|
|