

Next: The Interaction of Learning
Up: A Framework for GBML
Previous: Classifying GBML Systems by
Contents
Subsections
2.2 Classifying GBML Systems Algorithmically
In the Pittsburgh (Pitt) approach one chromosome encodes one solution.
We assume fitness is assigned to chromosomes, so in Pitt systems it is
assigned to solutions. This leaves a credit assignment problem: how
did the chromosome's component genes contribute to the observed
fitness of the chromosome? This is left to evolution as this is what
EAs are designed to deal with.
In the Michigan approach one solution is (typically) represented by
many chromosomes and so fitness is assigned to partial solutions.
Credit assignment differs from the Pitt case as chromosomes not only
compete for reproduction but may also complement and cooperate with
each other. This gives rise to the issues of how to encourage
cooperation, complementarity, and coverage of inputs, all of which makes
designing an effective fitness function more complex than in Pitt
systems. In Michigan systems the credit assignment problem is how to
measure a chromosome's contributions to the overall solution, as
reflected in the various aspects of fitness just mentioned. To sum up
the difficulty in Michigan systems: the best set of chromosomes may
not be the set of best (i.e. fittest) chromosomes
[95].
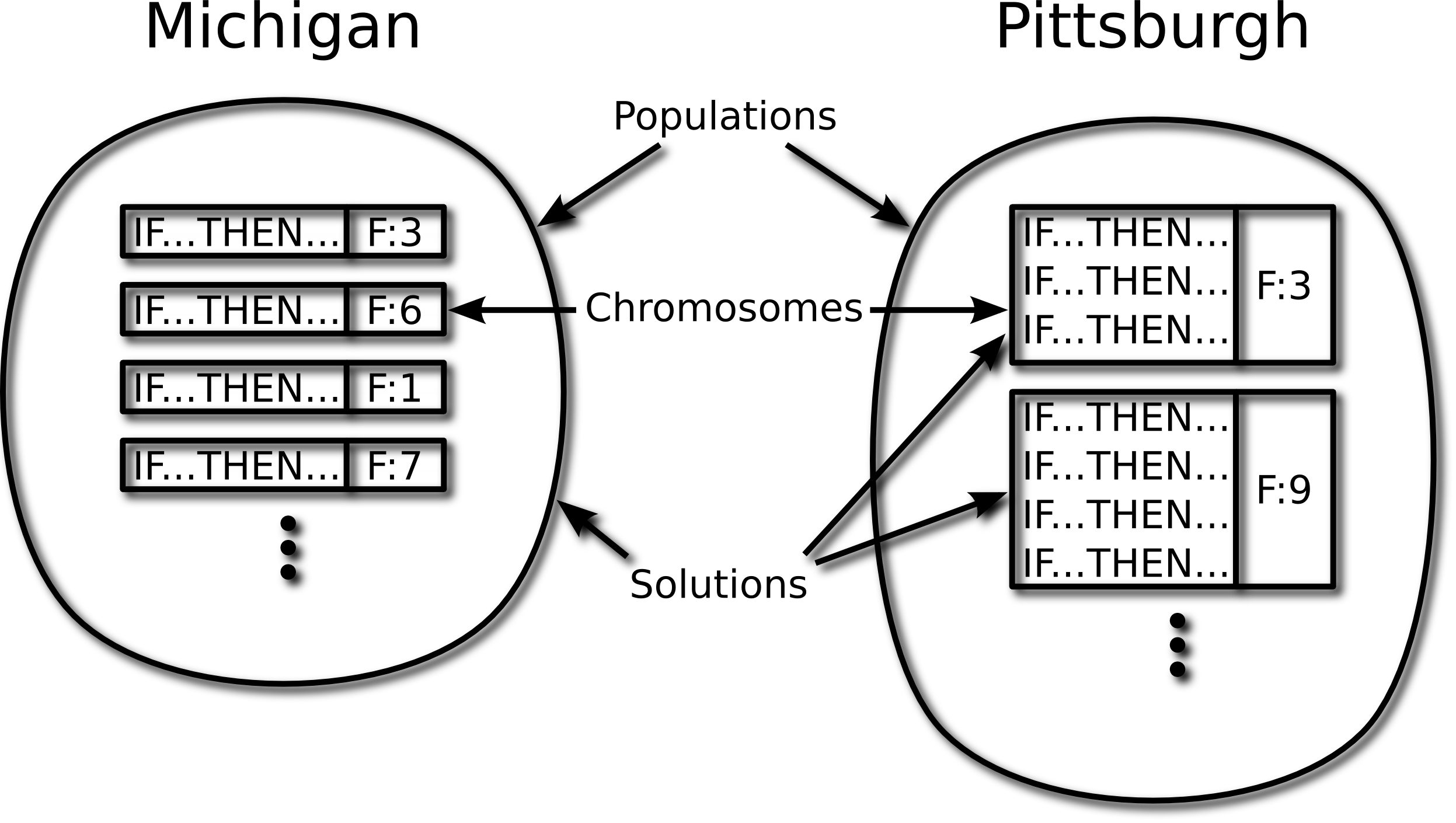
To illustrate, figure 2 depicts the
representation used by Pitt and Michigan versions of the rule-based
systems called Learning Classifier Systems (see §3.5). In
a Pittsburgh LCS a chromosome is a variable-length set of rules
while in a Michigan LCS a chromosome is a single fixed-length rule.
Figure 2:
Michigan and Pittsburgh rule-based systems compared. The
F: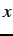 associated with each chromosome indicates its fitness
associated with each chromosome indicates its fitness
|
|
Although the Pittsburgh and Michigan approaches are generally
presented as two discrete cases some hybrids exist
e.g. [297].
Pittsburgh systems
(especially naive implementations) are slower, since they evolve more
complex structures and they assign credit at a less specific (and
hence less informative) level.1Additionally, since their chromosomes
are more complex so are their genetic operators. On the other hand
they face less complex credit assignment problems and are hence more
robust, that is, more likely to adapt successfully.
Michigan systems use a finer grain of credit assignment than the
Pittsburgh approach, which means bad partial solutions can be deleted
without restarting from scratch. This makes them more efficient and
also more suitable for incremental learning. However, credit
assignment is more complex in Michigan systems. Since the solution is
a set of chromosomes: i) the population must not converge fully,
and ii) as noted the best set of chromosomes may not be the set of
best chromosomes.
The two approaches also tend to be applied in different ways. Pitt
systems are typically used offline and are algorithm-driven; the main
loop processes each chromosome in turn and seeks out data to evaluate
them (which is how a standard GA works, although fitness evaluation is
typically simpler in a GA). In contrast, Michigan systems are typically used
online and are data-driven; the main loop processes each data input in
turn and seeks out applicable chromosomes (see figure
3). As a result Michigan systems are more
often used as learners (though not necessarily more often as
meta-learners) for reinforcement learning, which is almost always
on-line. The Michigan approach has mainly been used with LCS.
See
[111,141,297,96,154]
for comparison of the approaches.
Figure 3:
A basic Michigan algorithm
 |
IRL is a variation on the
Michigan approach in which, as usual, one solution is represented by
many chromosomes, but only the single best chromosome is selected
after each run, which alters the co-evolutionary dynamics of the
system. The output of multiple runs is combined to produce the
solution. The approach originated with SIA (Supervised Inductive
Algorithm) [287,190], a supervised genetic rule
learner.
Genetic Cooperative-Competitive Learning
GCCL is another Michigan approach in which on each generation is
ranked by fitness and a coverage-based filter then allocates
inputs to the first rule which correctly covers them. Inputs are only
allocated to one rule per generation and rules which have no inputs
allocated die at the end of a generation. The remaining rules' collective
accuracy is compared to the previous best generation, which is stored
offline. If the new generation is more accurate (or the same but has
fewer rules) it replaces the previous best. Examples include COGIN
[111,112], REGAL [105], and
LOGENPRO [312].



Next: The Interaction of Learning
Up: A Framework for GBML
Previous: Classifying GBML Systems by
Contents
T Kovacs
2011-03-12